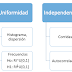
VARIABLES:
'ATENCION', 'SEXO', 'TIPO', 'ESTADO', 'TIPO_RECUPERACION', 'TID'
DATA: INS (National Health Institution) Colombia.
CODE:
from pandas import Series, DataFrame
import pandas as pd
import numpy as np
import matplotlib.pylab as plt
from sklearn.model_selection import train_test_split
from sklearn import preprocessing
from sklearn.cluster import KMeans
import pandas as pd
data=pd.read_csv('COVID/COVID_coursera_v4.csv',encoding='latin1', delimiter=';')
data.head()
data.columns = map(str.upper, data.columns)
list(data.columns)
cluster=data[['ATENCION', 'SEXO', 'TIPO', 'ESTADO', 'TIPO_RECUPERACION', 'TID']]
cluster.describe()
clustervar=cluster.copy()
from sklearn import preprocessing
clustervar['ATENCION']=preprocessing.scale(clustervar['ATENCION'].astype('float64'))
clustervar['SEXO']=preprocessing.scale(clustervar['SEXO'].astype('float64'))
clustervar['TIPO']=preprocessing.scale(clustervar['TIPO'].astype('float64'))
clustervar['ESTADO']=preprocessing.scale(clustervar['ESTADO'].astype('float64'))
clustervar['TIPO_RECUPERACION']=preprocessing.scale(clustervar['TIPO_RECUPERACION'].astype('float64'))
clustervar['TID']=preprocessing.scale(clustervar['TID'].astype('float64'))
clustervar.head()
clus_train, clus_test = train_test_split(clustervar, test_size=.3, random_state=123)
from scipy.spatial.distance import cdist
clusters=range(1,10)
meandist=[]
for k in clusters:
model=KMeans(n_clusters=k)
model.fit(clus_train)
clusassign=model.predict(clus_train)
meandist.append(sum(np.min(cdist(clus_train, model.cluster_centers_, 'euclidean'), axis=1))
/ clus_train.shape[0])
plt.plot(clusters, meandist)
plt.xlabel('Number of clusters')
plt.ylabel('Average distance')
plt.title('Selecting k with the Elbow Method')
from sklearn.decomposition import PCA
model3=KMeans(n_clusters=3)
model3.fit(clus_train)
clusassign=model3.predict(clus_train)
pca_2 = PCA(2)
plot_columns = pca_2.fit_transform(clus_train)
plt.scatter(x=plot_columns[:,0], y=plot_columns[:,1], c=model3.labels_,)
plt.xlabel('Canonical variable 1')
plt.ylabel('Canonical variable 2')
plt.title('Scatterplot of Canonical Variables for 5 Clusters')
plt.show()
clus_train.reset_index(level=0, inplace=True)
cluslist=list(clus_train['index'])
labels=list(model3.labels_)
newlist=dict(zip(cluslist, labels))
newlist
newclus=DataFrame.from_dict(newlist, orient='index')
newclus
newclus.columns = ['cluster']
newclus.reset_index(level=0, inplace=True)
merged_train=pd.merge(clus_train, newclus, on='index')
merged_train.head(n=100)
merged_train.cluster.value_counts()
clustergrp = merged_train.groupby('cluster').mean()
print ("Clustering variable means by cluster")
print(clustergrp)
edad_data=data['EDAD']
edad_train, edad_test = train_test_split(edad_data, test_size=.3, random_state=123)
edad_train1=pd.DataFrame(edad_train)
edad_train1.reset_index(level=0, inplace=True)
merged_train_all=pd.merge(edad_train1, merged_train, on='index')
sub1 = merged_train_all[['EDAD', 'cluster']].dropna()
import statsmodels.formula.api as smf
import statsmodels.stats.multicomp as multi
gpamod = smf.ols(formula='EDAD ~ C(cluster)', data=sub1).fit()
print (gpamod.summary())
print ('means for EDAD by cluster')
m1= sub1.groupby('cluster').mean()
print (m1)
print ('standard deviations for GPA by cluster')
m2= sub1.groupby('cluster').std()
print (m2)
mc1 = multi.MultiComparison(sub1['EDAD'], sub1['cluster'])
res1 = mc1.tukeyhsd()
print(res1.summary())
RESULTS:
A k-means cluster analysis was conducted to identify underlying subgroups of people based on their similarity of responses on 6 variables that represent characteristics that could have an impact on EDAD (age). Clustering variables included three binary variables measuring whether or not the person is male or female and if it is recovered or deceased and if for his recovery the person used PCR or just time, as well as quantitative variables measuring Time from symptoms appeared to diagnosis, a scale measuring ESTADO (Status), and scales measuring TIPO. All clustering variables were standardized to have a mean of 0 and a standard deviation of 1.
Data were randomly split into a training set that included 70% of the observations (N=29409) and a test set that included 30% of the observations (N=12604). A series of k-means cluster analyses were conducted on the training data specifying k=1-9 clusters, using Euclidean distance. The variance in the clustering variables that was accounted for by the clusters (r-square) was plotted for each of the nine cluster solutions in an elbow curve to provide guidance for choosing the number of clusters to interpret.
Figure 1. Elbow curve of r-square values for the nine cluster solutions
The elbow curve was inconclusive, suggesting that the 3, 5 and 8-cluster solutions might be interpreted. The results below are for an interpretation of the 3-cluster solution.
Canonical discriminant analyses was used to reduce the 6 clustering variable down a few variables that accounted for most of the variance in the clustering variables. A scatterplot of the first two canonical variables by cluster (Figure 2 shown below) indicated that the observations in clusters 1 and 2 were densely packed with relatively low within cluster variance, and did not overlap very much with the other clusters. Cluster 3 was generally distinct, but the observations had greater spread suggesting higher within cluster variance. Observations in cluster 3 were spread out more than the other clusters, showing high within cluster variance. The results of this plot suggest that the best cluster solution may have more than 3 clusters, so it will be especially important to also evaluate the cluster solutions with fewer than 5 clusters.
Figure 2. Plot of the first two canonical variables for the clustering variables by cluster.
The means on the clustering variables showed that, compared to the other clusters, EDAD in cluster 0 had moderate levels on the clustering variables. They had a relatively high likelihood of ATENCION and TID, but moderate levels of SEXO and TIPO. They also appeared to have fairly low levels of ESTADO, cluster 1 had higher levels on ESTADO variables compared to cluster 0. On the other hand, cluster 2 had the highest likelihood in TIPO, and the lowest levels of SEXO and TID.
In order to externally validate the clusters, an Analysis of Variance (ANOVA) was conducting to test for significant differences between the clusters on grade point average (GPA). A tukey test was used for post hoc comparisons between the clusters. Results indicated significant differences between the clusters on GPA (F=175.5, p<.0001). The tukey post hoc comparisons showed significant differences between clusters on GPA. People in cluster 1 had the highest GPA (mean=40.37, sd=19.41), and cluster 2 had the lowest GPA (mean=35.10, sd=17.08).